About
From small molecules to complex organelles
The targeted degradation of proteins is of utmost importance for the well-being of our cells and thus for the entire organism. Damaged proteins must be recognized and removed to prevent collateral damage. In addition, proteins must be removed in a targeted manner in order to allow the activation, but also the deactivation of cellular signal transduction pathways. Defects in the targeted degradation of proteins can result in diseases such as cancer and neurodegeneration.
Our Special Research Program, SFB F79, is funded by the Austrian Science Fund (FWF) with contributions by the German Research Foundation (DFG). The Program grew out of the Ubiquitin Club, a group of scientists at the Vienna BioCenter fascinated by the ubiquitin protein and its many cellular functions. In the expanded collaborative research program scientists from Max Perutz Labs, University of Vienna, Medical University of Vienna, CeMM, GMI, IMBA and IMP from Vienna as well as MDC in Berlin join forces to discover mechanism of targeted protein degradation, and how this process can be manipulated.
Ubiquitin-proteasome system
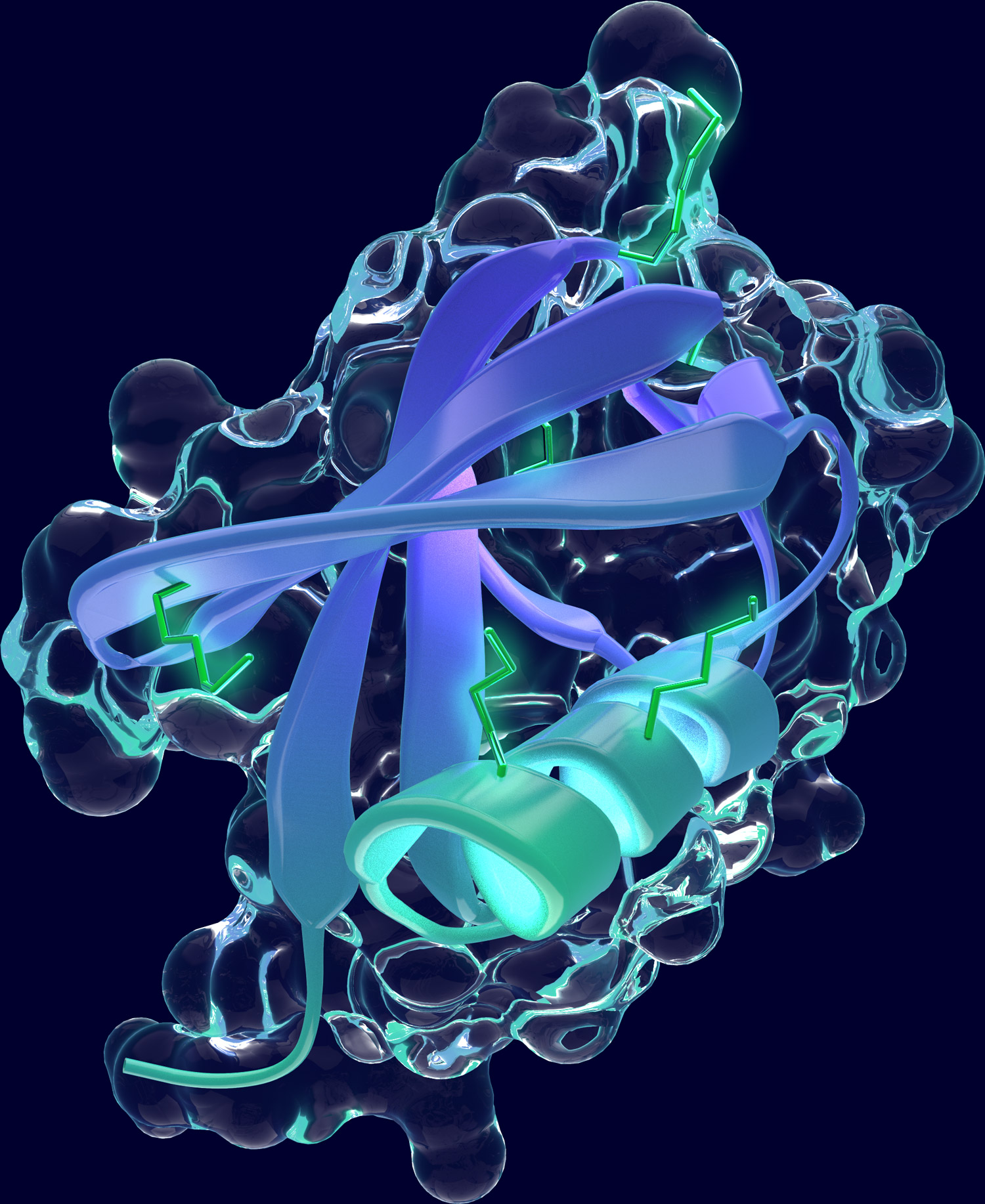
The ubiquitin-proteasome system (UPS) is one the major protein degradation pathways. In the UPS, proteins destined for degradation are marked by the attachment of the small protein ubiquitin, a covalent posttranslational protein modification catalyzed by the cascade of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin-ligase (E3). Ubiquitination serves a tag for protein substrates to be recognized by the proteasome, a large and highly organized multicatalytic protease complex, where substrates are unfolded and degraded. Once attached to a protein, ubiquitin itself can be subject to further modifications and can serve as substrate for further ubiquitin conjugation at different lysine residues. Thus, substrates can be tagged by a single ubiquitin or with ubiquitin chains of diverse length and branches.
Autophagy
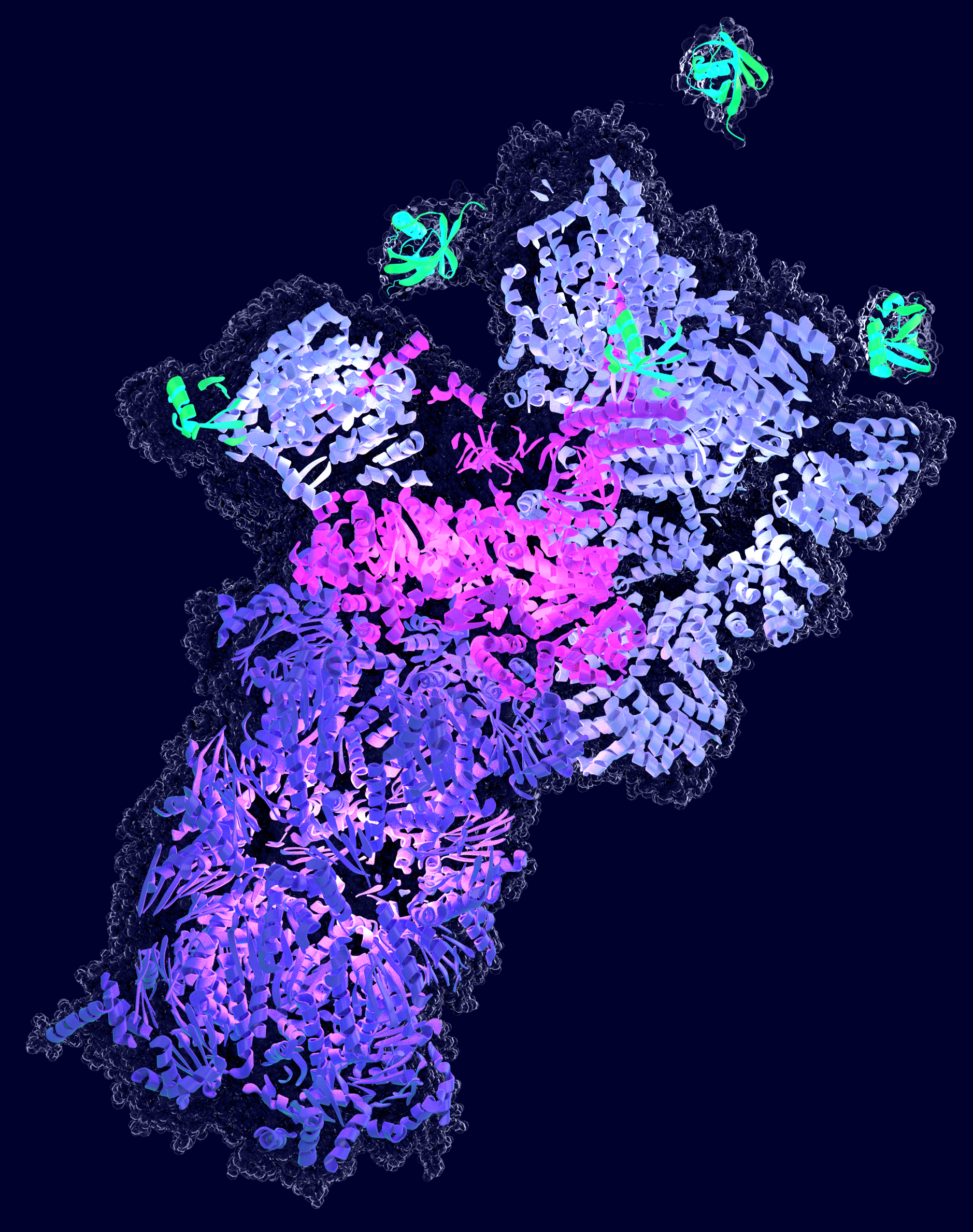
The second major degradation system in our cells is autophagy, which delivers proteins into the lysosome for proteolysis. During one form of autophagy, cytoplasmic material is engulfed by double-membrane organelles, called autophagosomes, that subsequently fuse with lysosomes, wherein their content is degraded. Selective autophagy involves the binding of the “cargo” by a cargo receptor and two of the major cargo receptors for autophagy recognize ubiquitin as a degradation signal. Thus, selective autophagy and the UPS perceive the same protein modification to select their target proteins.
How such ubiquitinated proteins destined for degradation are channeled towards either the UPS or to autophagy, is addressed by our research network. While both of these two systems act in parallel in the cytoplasm, their crosstalk is unclear. On the other hand, in the nucleoplasm, autophagy is generally not known to act, but the long-term fate of nuclear aggregated proteins that cannot be dealt with by the UPS is unknown and subject of investigation within our network.
Artificial induction of protein degradation by small molecules
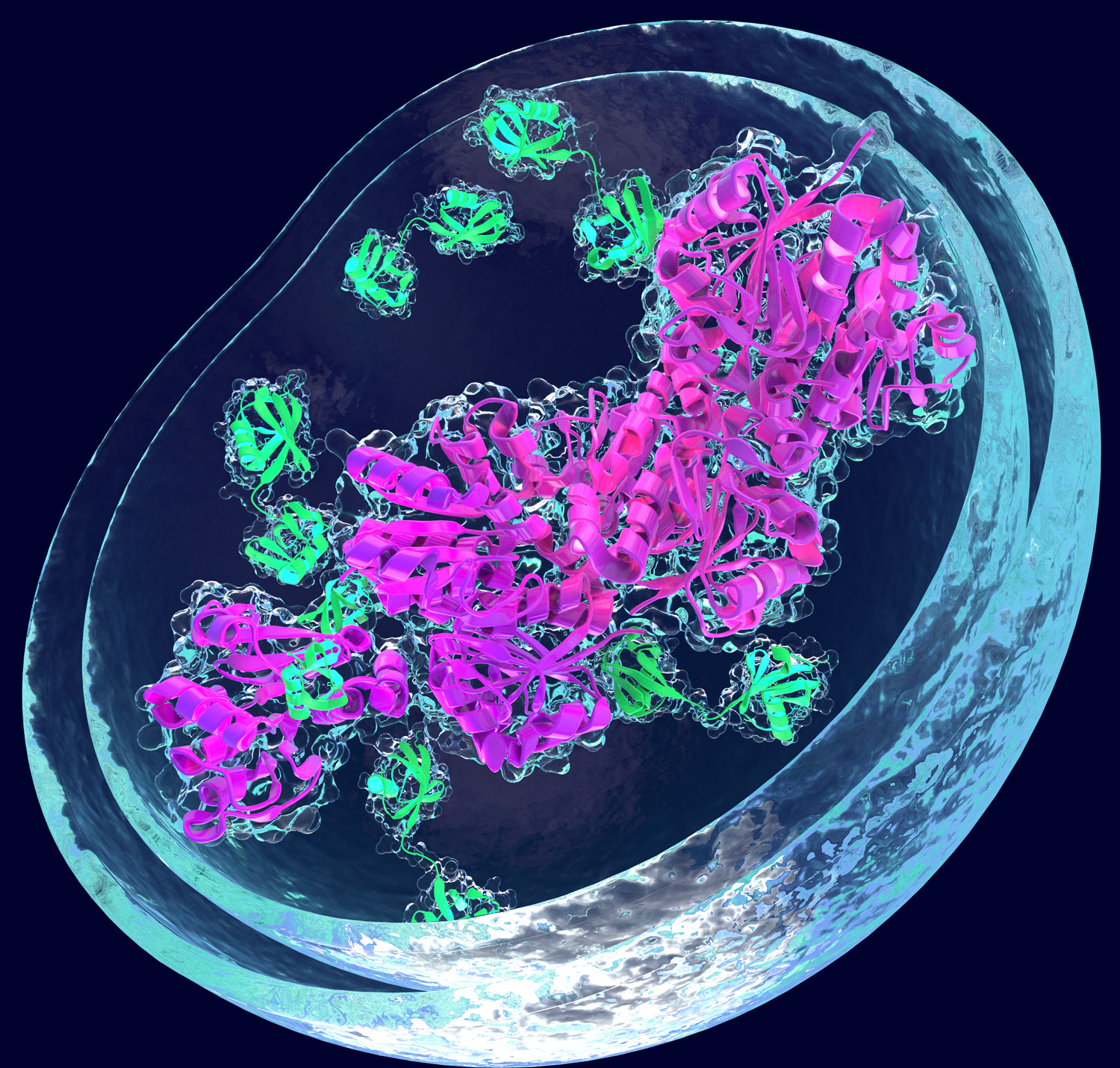
A third major focus of our research program is how the degradation systems can be reprogrammed to induce the degradation of otherwise stable and harmful proteins by using small molecules. Reprogramming approaches have great promises in the treatment of a plethora of diseases. Within our consortium, we are investigating how small molecules can be used to chemically reprogram the association of distinct E3 ligases and proteins of interest, enabling the targeted proteolysis of selected proteins in a spatially and temporally controlled manner. Small molecules can act as “molecular glues” that can induce ternary complex formation between a target protein and an E3 ligase. In addition, heterobifunctional molecules known as PROTACs (proteolysis targeting chimeras) link a desired target protein to a E3 ubiquitin ligase. Extending these approaches is among the aims of our research.
Scientific Advisory Board
-
Stanford University
Judith Frydman
Judith Frydman is the Donald Kennedy Chair in Humanities and Sciences and a professor in the Departments of Biology and Genetics at Stanford University. Originally from Buenos Aires, Argentina, where she majored in Chemistry and received her PhD in Biochemistry from the University of Buenos Aires, she carried out her postdoctoral training with Ulrich Hartl at the Sloan Kettering Institute in New York, where she had major contributions to the field of cellular protein folding. The Frydman lab has made seminal contributions to understand how eukaryotic cells manage their proteome and maintain protein homeostasis. Her studies have important applications to ameliorate aging, neurodegenerative diseases and other misfolding-linked maladies and enable strategies for the treatment of infectious disease and neurodegenerative diseases.
-
MRC Laboratory of Molecular Biology
Ramanujan Hegde
Ramanujan Hegde is a group leader and Joint Head of the Cell Biology Division at the MRC Laboratory of Molecular Biology in Cambridge, UK. Research in the Hegde lab focuses on the biogenesis of membrane proteins and the various quality control pathways that deal with failed protein maturation. Ramanujan Hegde is an elected member of EMBO and a Fellow of the Royal Society.
-
Boston Children's Hospital & Harvard Medical School
Hidde Ploegh
Hidde Ploegh obtained his Master of Science degree in biology and chemistry in 1977 from the University of Groningen, and pursued his Ph.D. studies in the lab of Jack Strominger. He received a doctorate from the University of Leiden. He has worked at the University of Cologne, the Netherlands Cancer Institute, Utrecht University, and Harvard Medical School, and is a member of the Whitehead Institute. Ploegh is member of EMBO and of the National Academy of Sciences.
-
University of Liverpool
Sylvie Urbé
Sylvie Urbé is a cell biologist from Luxembourg who studied at the University of Heidelberg, Germany and was trained at the European Molecular Biology Laboratory (EMBL; ultrastructural localisation and function of Rab GTPases) and Imperial Cancer Research Fund in London, UK (cell-free assays of secretory granule maturation). She was awarded successive Wellcome Trust and Cancer Research UK Research Fellowships to study the endo-lysosomal protein sorting machinery and associated DUBs at the University of Liverpool where she now holds a Professorship. Her current interests still have a membrane traffic and organelle biology flavour, with ubiquitin-modifying enzymes (DUBs and E3 ligases) a constant theme in ongoing projects.