Group Karagöz
Dissecting mechanisms regulating Stress Granule assembly and dissolution
Group Leader
Elif Karagöz
Elif Karagöz majored Molecular Biology with a minor in Chemistry at the Middle East Technical University in Turkey. After completing her PhD at Utrecht University with Stefan Rüdiger, she did a postdoc at Peter Walter’s lab at the University of California San Francisco. Since 2019 she is Group Leader at the Max Perutz Labs Vienna.
- Institute Max Perutz Labs, Medical University of Vienna
- Phone +43 1 4277 61635
- Mail elif.karagoez@univie.ac.at
- Web https://www.maxperutzlabs.ac.at/karagoez
Projects within consortium
To cope with protein-folding stress, cells transiently shut down protein translation. Under these conditions, mRNAs and select RNA-binding proteins assemble into dynamic cytoplasmic complexes called stress granules (SG) that form through a liquid-liquid phase separation process. Perturbed SG turnover contributes to protein aggregation in aging. Protein quality control machineries regulate SG disassembly and clearance. Yet, their precise role and mechanism of action is poorly understood. We aim to dissect how SG components are recognized and processed by the quality control machinery to hand over to UPS and autophagy using complementary approaches.
Stress granules are membraneless organelles that regulate mRNA stability during stress induced translational shutdown (Anderson and Kedersha, 2009; Buchan and Parker, 2009). They are dynamic condensates, which are formed by a liquid-liquid phase separation process through the interaction of RNAs and several RBPs with intrinsically disordered domains. Perturbed SG turnover contributes to protein aggregation due to high aggregation propensity of the intrinsically disordered proteome found in SGs. Recent evidence suggests that the protein quality control and degradation pathways play a crucial role in regulating SG (dis-)assembly to maintain SG composition, integrity, and dynamics (Gamerdinger et al, 2011; Buchan et al, 2013; Seguin et al, 2014). Select chaperones and the triple ATPase VCP machinery co-localize to SGs and regulate SG disassembly and clearance through the action of UPS and autophagy machinery (Kwon et al, 2007; Buchan et al, 2013; Seguin et al, 2014). However, how the quality control machinery recognizes SG proteome and their precise role and the mechanism of action is poorly understood.
To fill the gap in our knowledge, we will combine cell biology, microscopy and in vitro reconstitution approaches to reveal how SGs are recognized by the molecular chaperones and the VCP and handed over to either UPS or autophagy machineries for degradation. We anticipate that the overlapping topics of interest and complementary technical expertise in this consortium will provide a strong basis to answer these fundamental questions in cell biology.
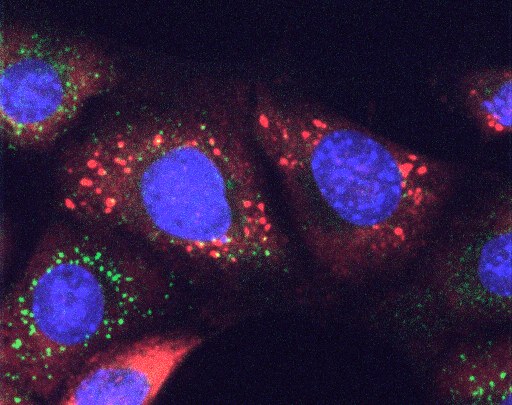
Stress Granules (red) are formed after stress induction by drugs that impair protein folding in the ER. Mouse embryonic fibroblasts stained for the SG marker G3BP2 (red), the ER stress sensor IRE1 (green) and the nucleus (blue).
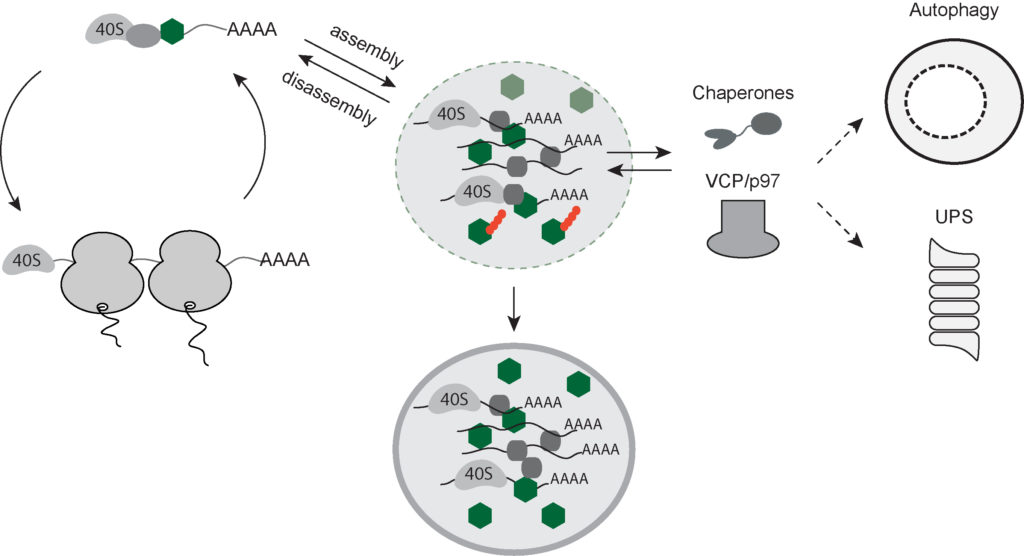
Current model for the SG (dis-)assembly.
Project members
-
PhD Student
Aleksandra Anisimova
SFB Member
-
PhD Student
Harald Hornegger
SFB Member
Targeted Protein Degradation related publications by Group Karagöz
- 2025 The coordinated action of UFMylation and ribosome-associated quality control pathway clears arrested nascent chains at the endoplasmic reticulum bioRxiv Go to publication →
- 2024 IGF2BP1 phosphorylation in the disordered linkers regulates ribonucleoprotein condensate formation and RNA metabolism Nature Communications Go to publication →
- 2024 Disordered regions in the IRE1α ER lumenal domain mediate its stress-induced clustering EMBO Journal Go to publication →
- 2024 Guardian ubiquitin E3 ligases target cancer-associated APOBEC3 deaminases for degradation to promote human genome integrity bioRxiv Go to publication →
- 2024 Endoplasmic reticulum: Monitoring and maintaining protein and membrane homeostasis in the endoplasmic reticulum by the unfolded protein response Int J Biochem Cell Biol Go to publication →
- 2023 Optimized infrared photoactivatable ribonucleoside enhanced crosslinking and immunoprecipitation (IR-PAR-CLIP) protocol identifies novel IGF2BP3-interacting RNAs in colon cancer cells. RNA Go to publication →
- 2023 Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and C53-mediated autophagy EMBO Journal Go to publication →
- 2018 The unfolded protein response and endoplasmic reticulum protein targeting machineries converge on the stress sensor IRE1 eLife Go to publication →
- 2017 An unfolded protein-induced conformational switch activates mammalian IRE1 eLife Go to publication →
- 2014 Hsp90-Tau complex reveals molecular basis for specificity in chaperone action Cell Go to publication →