Group Bachmair
N-degron pathways in plants
Group Leader
Andreas Bachmair
After his PhD in Biochemistry in Vienna, Andreas Bachmair did postdoctoral research at the MIT in Cambridge, USA, and at the MPI for Plant Breeding Research in Germany. 1991-2002 he was Assistant Professor at the University of Vienna, 2002-2008 Group Leader at the MPI for Plant Breeding Research, and in 2009 back to the University of Vienna, first as Associate Professor and since 2019 full Professor at the Max Perutz Labs.
- Institute Max Perutz Labs, Univ. of Vienna
- Phone +43 1 4277 74811
- Mail andreas.bachmair@univie.ac.at
- Web https://www.maxperutzlabs.ac.at/bachmair
Projects within consortium
The amino terminus of a protein is a determinant of its half-life. Using the model plant Arabidopsis thaliana, we investigate how proteins are turned over that have N-terminal amino acids with bulky side chain. We are interested in pathway components and in signal transduction cascades that depend on these pathways. Particular focus lies on N-terminal Leu, because for these proteins, the degradation path is currently uncharacterized.
The amino terminus is an important determinant of a protein´s half-life. This pertains not only to primary translation products, but even more to products of proteolytic processing, generated by over five hundred proteases encoded in every metazoan genome. Many of the neo-N-termini generated by these latter enzymes are signals that shorten the protein half-life, channeling affected proteins into degradation routes called N-degron pathways. Unacetylated first amino acids can act as degradation signals if they have either a hydrophobic, or a basic side chain. Degradation of proteins with basic first residue is mechanistically similar in animals and in plants, and participates in oxygen sensing. In contrast, turnover of proteins with hydrophobic N-terminal residues uses pathways that differ between animals and plants. Of particular interest to us is the degradation route for plant proteins with aliphatic hydrophobic amino-terminal residues such as Leu, because it is still uncharacterized. Our preliminary data suggest that both the ubiquitin-proteasome system, and autophagy participate in degradation. We are studying the pathway components using model substrates and a broad array of genetic, biochemical and cell biology methods. We also want to identify signal transduction cascades that utilize these pathways in stress response and development.
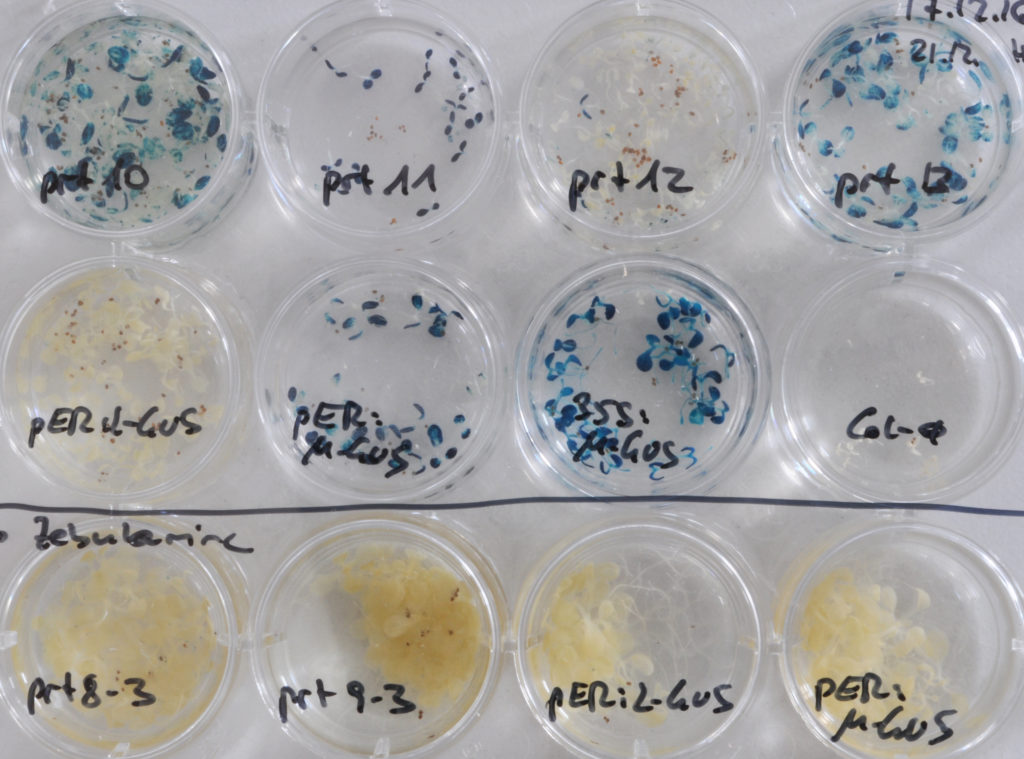
Accumulation of a GUS protein with N-terminal Leu in prt (proteolysis) mutant seedlings can be visualized by increased conversion of a chromogenic substrate into a blue precipitate.
Project members
-
Postdoc
Carolina Saad
SFB Member
-
former Postdoc
Nikola Winter
(until 2024-02)
Targeted Protein Degradation related publications by Group Bachmair
- 2025 The UBR domain of plant Ubr1 homolog PRT6 accommodates basic and hydrophobic amino termini for substrate recognition Journal of Molecular Biology Go to publication →
- 2024 ATG8ylation of vacuolar membrane protects plants against cell wall damage bioRxiv Go to publication →
- 2024 BIG enhances Arg/N-degron pathway-mediated protein degradation to regulate Arabidopsis hypoxia responses and suberin deposition Plant Cell Go to publication →
- 2023 Analysis of higher plant N-degron pathway components and substrates via expression in S. cerevisiae Methods in Enzymology Go to publication →
- 2023 SUMO Conjugation and SUMO Chain Formation by Plant Enzymes Methods in Molecular Biology Go to publication →
- 2022 Transcriptome, metabolome and suppressor analysis reveal an essential role for the ubiquitin-proteasome system in seedling chloroplast development BMC Plant Biology Go to publication →
- 2022 A yeast-based functional assay to study plant N-degron – N-recognin interactions Frontiers in Plant Science Go to publication →
- 2021 Cellular control of protein turnover via modification of the amino terminus International Journal of Molecular Sciences Go to publication →
- 2019 The Scope, Functions, and Dynamics of Posttranslational Protein Modifications Annu. Rev. Plant Biol. Go to publication →
- 2019 Distinct branches of the N-end rule pathway modulate the plant immune response New Phytologist Go to publication →
- 2018 SUMO chain formation relies on the amino-terminal region of SUMO conjugating enzyme and has dedicated substrates in plants Biochemical J. Go to publication →
- 2014 The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions Trends in Cell Biol. Go to publication →
- 2014 Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors Mol. Cell Go to publication →